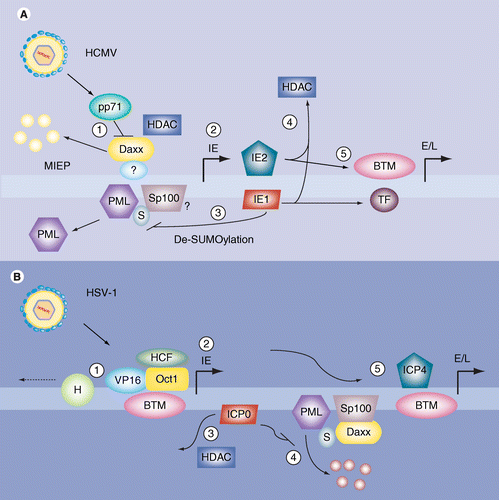
Promyelocytic leukemia-nuclear body proteins: herpesvirus enemies, accomplices, or both? | Future Virology

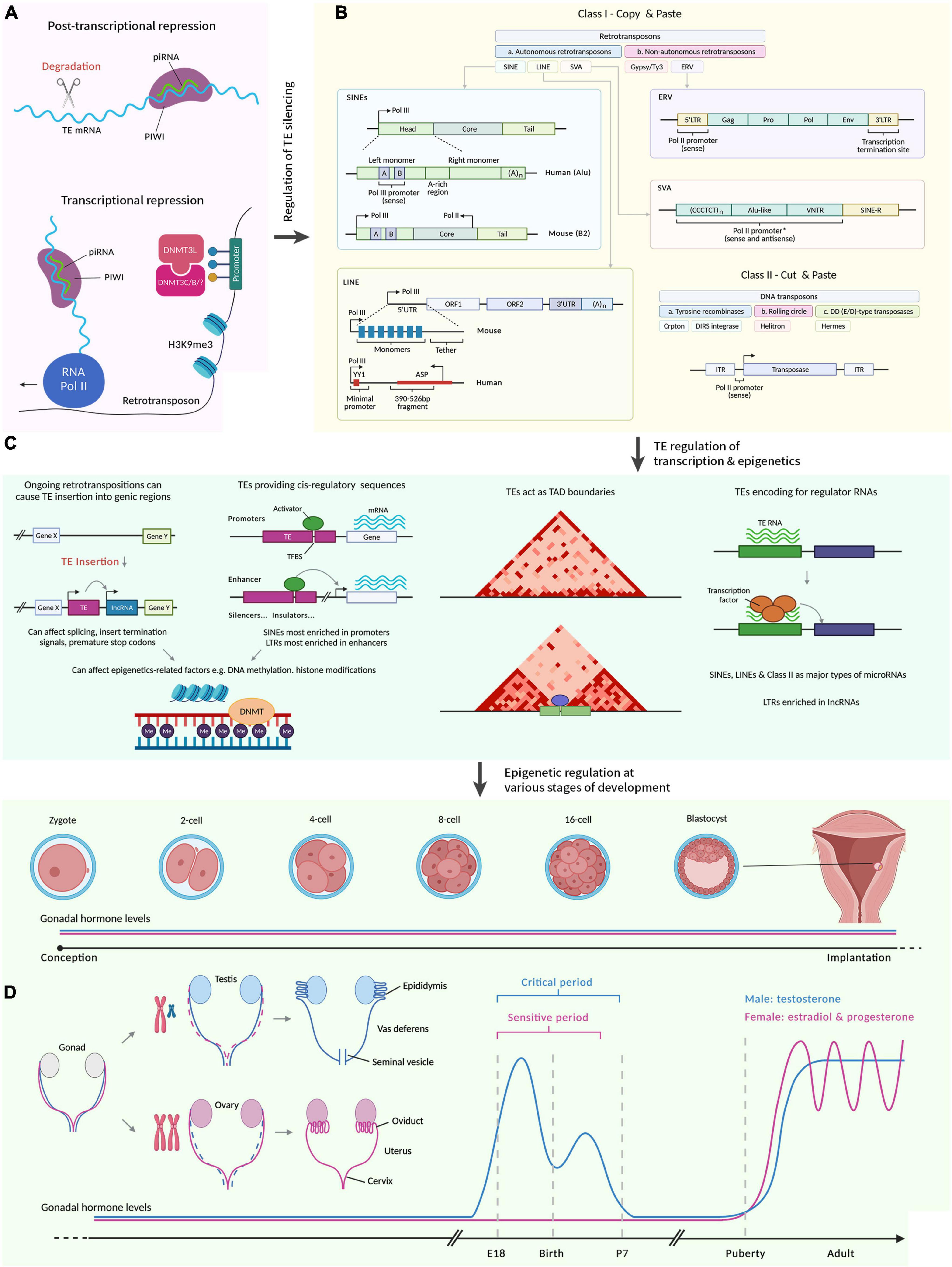
Relationship between DNA methylation and H3K9me3 at ZFP57-binding sites... | Download Scientific Diagram
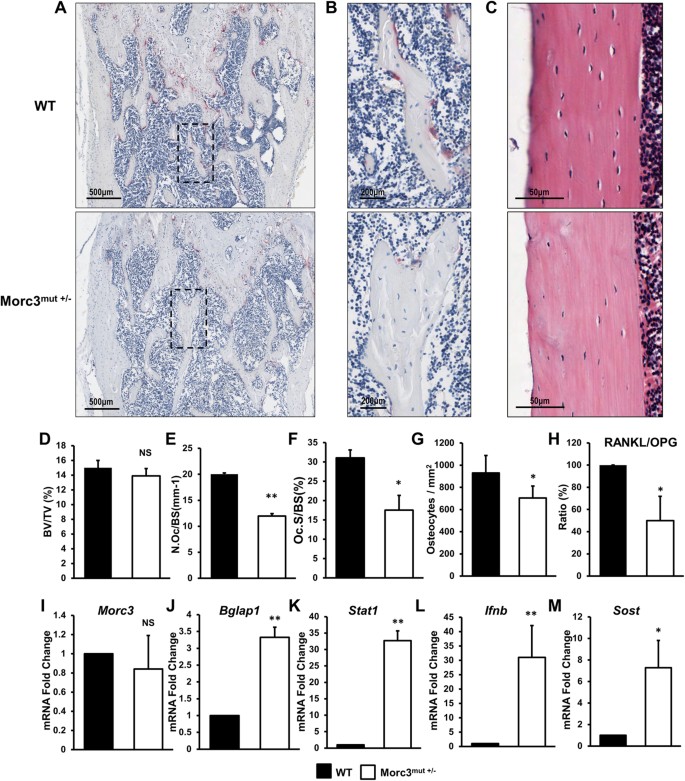
Morc3 mutant mice exhibit reduced cortical area and thickness, accompanied by altered haematopoietic stem cells niche and bone cell differentiation | Scientific Reports

Frontiers | A Tale of Usurpation and Subversion: SUMO-Dependent Integrity of Promyelocytic Leukemia Nuclear Bodies at the Crossroad of Infection and Immunity
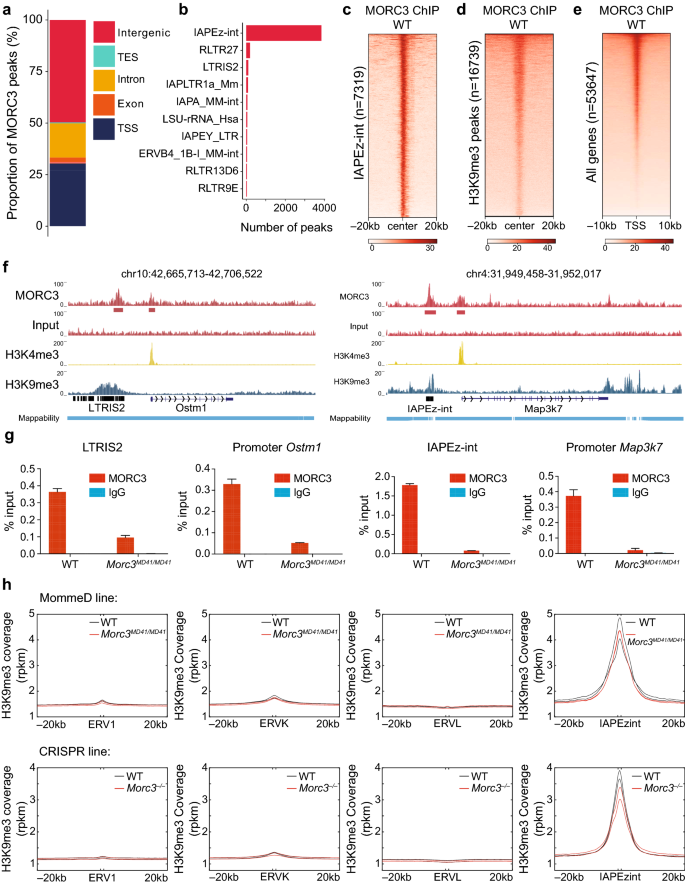
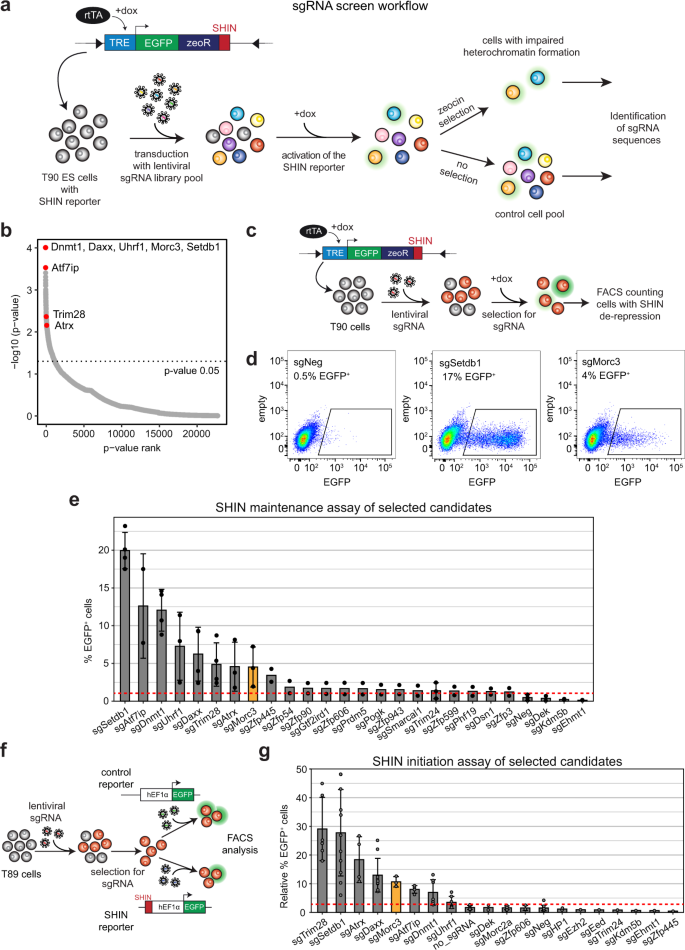
The role of MORC3 in silencing transposable elements in mouse embryonic stem cells | Epigenetics & Chromatin | Full Text
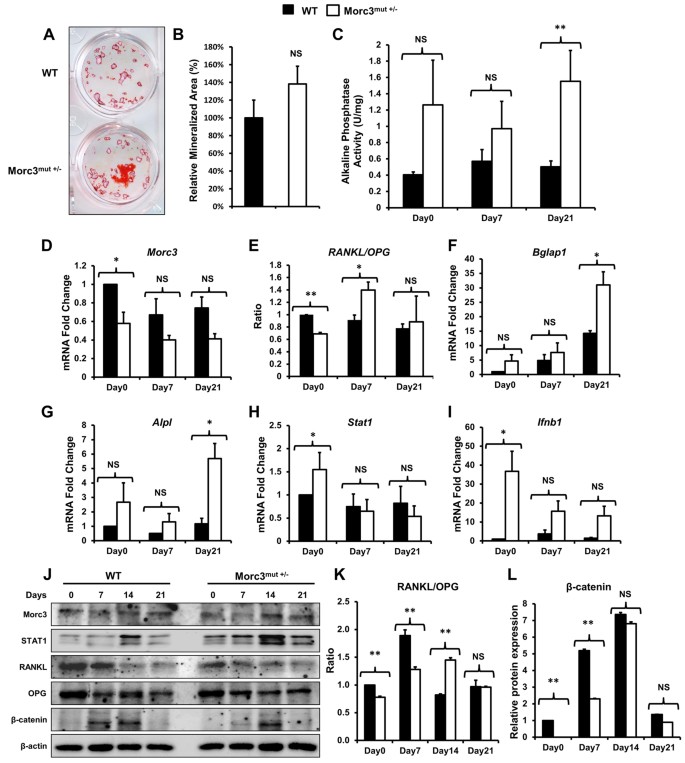
Morc3 mutant mice exhibit reduced cortical area and thickness, accompanied by altered haematopoietic stem cells niche and bone cell differentiation | Scientific Reports

Binding of the SARS-CoV-2 envelope E protein to human BRD4 is essential for infection - ScienceDirect
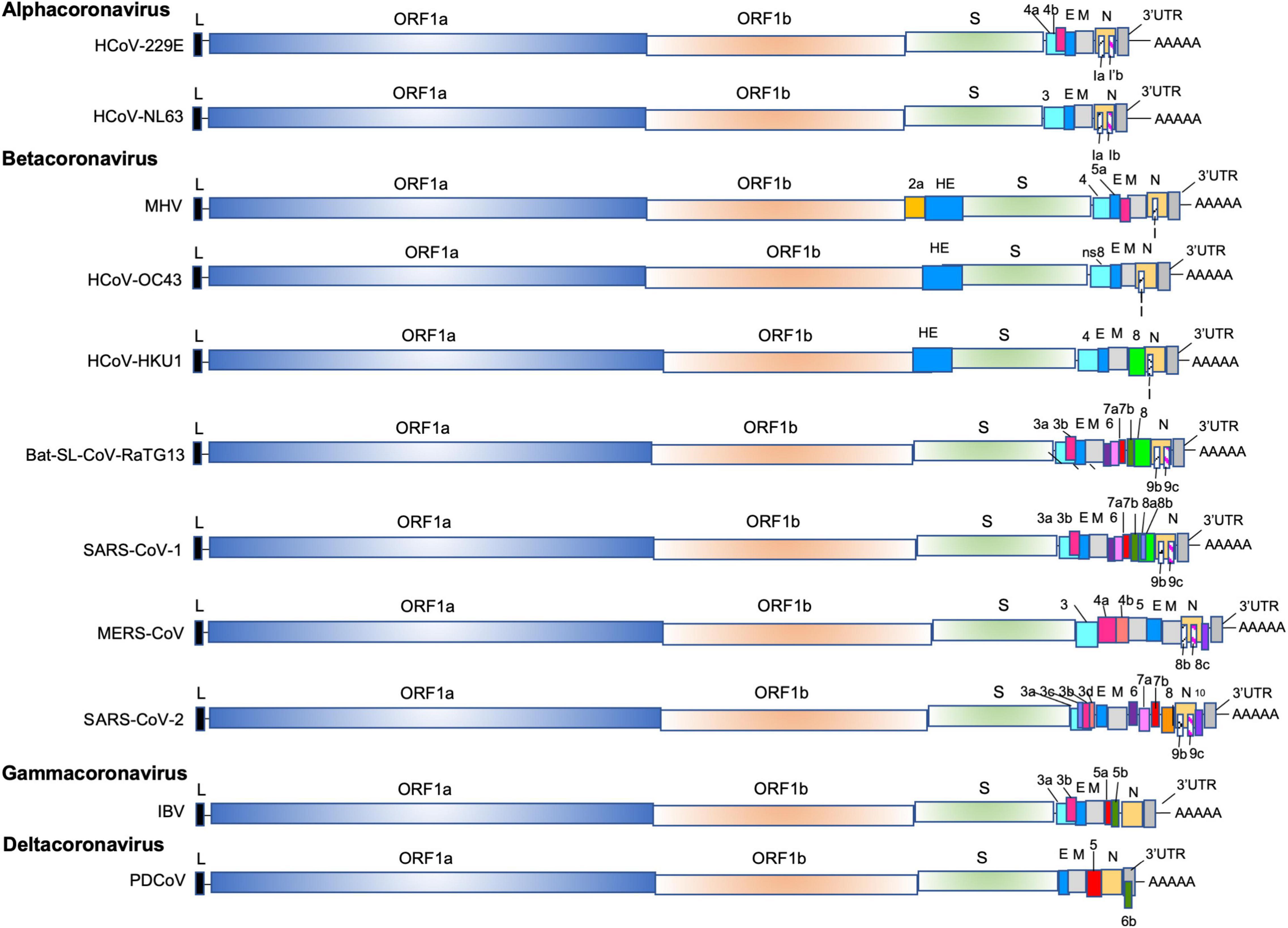
Frontiers | Coronavirus, the King Who Wanted More Than a Crown: From Common to the Highly Pathogenic SARS-CoV-2, Is the Key in the Accessory Genes?

The role of MORC3 in silencing transposable elements in mouse embryonic stem cells | Epigenetics & Chromatin | Full Text

Chemical Biology Approaches to Identify and Profile Interactors of Chromatin Modifications | ACS Chemical Biology

Uncovering the mechanistic basis for specific recognition of monomethylated H3K4 by the CW domain of Arabidopsis histone methyltransferase SDG8 - Journal of Biological Chemistry
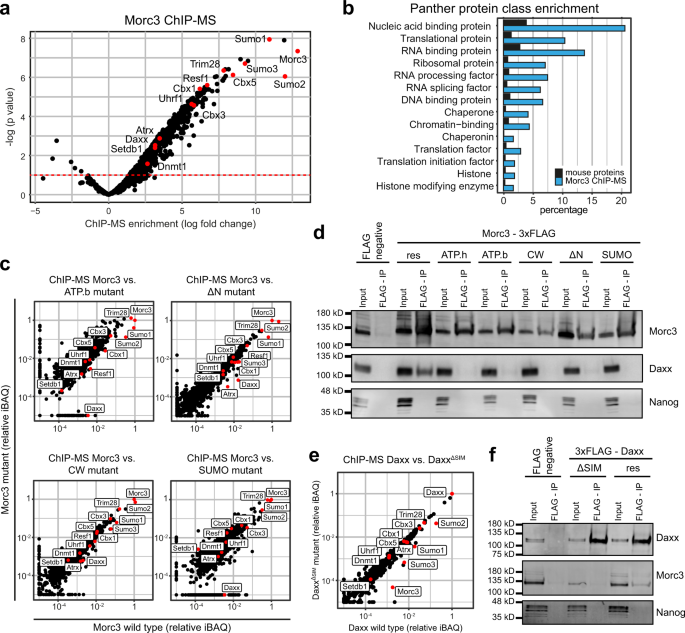
Morc3 silences endogenous retroviruses by enabling Daxx-mediated histone H3.3 incorporation | Nature Communications

Morc3 mutant mice exhibit reduced cortical area and thickness, accompanied by altered haematopoietic stem cells niche and bone cell differentiation | Scientific Reports
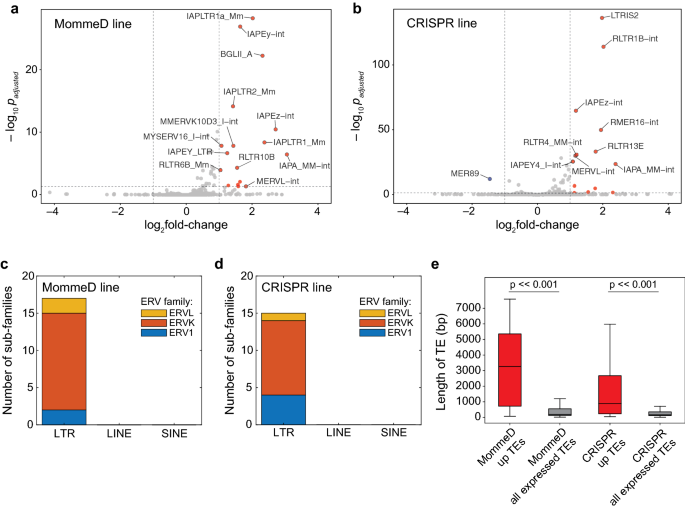
The role of MORC3 in silencing transposable elements in mouse embryonic stem cells | Epigenetics & Chromatin | Full Text

IJMS | Free Full-Text | Role of miRNAs in Human T Cell Leukemia Virus Type 1 Induced T Cell Leukemia: A Literature Review and Bioinformatics Approach | HTML